 |
| A descrição detalhada da camada externa do HIV, o capsídeo, vai ajudar no desenvolvimento de novas drogas que possam interromper sua formação, destruindo o poder de infecção do vírus. |
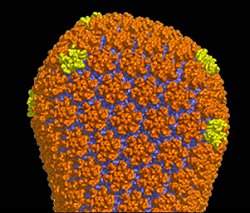
Capsídeo
Uma equipe de cientistas norte-americanos descreveu pela primeira vez a estrutura do conjunto de proteínas que fornece o material genético do vírus da imunodeficiência humana (HIV) para as células.
O trabalho é o resultado de estudos realizados nos últimos 10 anos, concentrados nas diferentes partes do recipiente em forma de cone do vírus, o capsídeo.
A última peça do quebra-cabeças foi descrita em um artigo publicado na revista Nature por cientistas da Universidade da Virgínia e do Instituto de Pesquisas Scripps.
"Este artigo é um verdadeiro marco," avalia o autor sênior do estudo, Dr. Mark Yeager.
A descrição detalhada do capsídeo do HIV vai fornecer um roteiro para o desenvolvimento de drogas que possam prejudicar a sua formação e, assim, prevenir a infecção pelo HIV.
Como o HIV se reproduz
O HIV se liga a receptores nas células humanas e depois injeta o capsídeo dentro delas. Uma vez dentro da célula, o capsídeo se desfaz, liberando o material genético do vírus.
Em seguida, o HIV sabota o maquinário celular para fazer muitas cópias de seus genes e proteínas.
Conforme novos vírus são feitos, o material genético é empacotado em capsídeos esféricos imaturos que o HIV usa para fugir da célula infectada.
Mas, antes que estes vírus recém-lançados possam infectar outras células, o capsídeo imaturo sofre uma dramática reorganização para formar a casca madura, em forma de cone.
Se a formação do capsídeo maduro for interrompido, o vírus não é mais infeccioso.
Assim, novas drogas dirigidas à formação do capsídeo poderão fornecer valiosas adições ao arsenal de drogas existentes contra o HIV.
Cristalografia
Para desenvolver drogas que interrompam a formação do capsídeo, no entanto, os cientistas primeiro precisam saber exatamente como ele é formado.
Isto não pode ser feito com a tecnologia tradicionalmente usada para descrição das estruturas detalhadas das moléculas biológicas, chamada cristalografia de raios X.
Diferentemente dos capsídeos em forma de cone de outros vírus, como os poliovírus, que têm uma estrutura rígida e simétrica, o capsídeo do HIV é flexível e pode adotar formas ligeiramente diferentes.
Parte dessa flexibilidade deve-se à proteína que compõe o capsídeo do HIV, a proteína CA, constituída por duas extremidades unidas por uma ponte "flexível".
Isto torna impossível crescer cristais do capsídeo completo do HIV para que a cristalografia possa ser usada.
Ciclo de vida do vírus
Os cientistas então partiram para a tática de dividir para conquistar, particionando o capsídeo do HIV em componentes menores e, em seguida, determinando suas respectivas estruturas.
Depois de descobrirem as estruturas atômicas dos hexâmeros e pentâmeros da proteína CA, Yeager e seus colegas agora conseguiram construir um modelo atômico completo do capsídeo do HIV maduro.
Os pesquisadores agora planejam aperfeiçoar o modelo utilizando sofisticados programas de computador para determinar a estabilidade da estrutura em diferentes regiões e identificar possíveis pontos "fracos" que possam ser alvo de novas drogas.
Eles vão também começar a estudar a estrutura do capsídeo imaturo para determinar como esta versão do capsídeo se metamorfoseia para a forma madura - uma etapa no ciclo de vida do vírus que permanece misteriosa.