- S. Bhaduri and J. G. PhillipsArticle first published online: 13 NOV 2009 | DOI: 10.1111/j.1863-2378.2009.01271.x
- J. H. McQuiston, M. A. Guerra, M. R. Watts, E. Lawaczeck, C. Levy, W. L. Nicholson, J. Adjemian and D. L. SwerdlowArticle first published online: 23 DEC 2009 | DOI: 10.1111/j.1863-2378.2009.01300.x
- C. S. Kyriakis, I. H. Brown, E. Foni, G. Kuntz-Simon, J. Maldonado, F. Madec, S. C. Essen, C. Chiapponi and K. Van ReethArticle first published online: 23 DEC 2009 | DOI: 10.1111/j.1863-2378.2009.01301.x
- B. Sibhat, B. Molla Zewde, A. Zerihun, A. Muckle, L. Cole, P. Boerlin, E. Wilkie, A. Perets, K. Mistry and W. A. GebreyesArticle first published online: 23 DEC 2009 | DOI: 10.1111/j.1863-2378.2009.01305.x
- L. Touihri, I. Zaouia, K. Elhili, K. Dellagi and C. BahloulArticle first published online: 23 DEC 2009 | DOI: 10.1111/j.1863-2378.2009.01306.x
- J. L. Jones, B. Anderson, J. Schulkin, M. E. Parise and M. L. EberhardArticle first published online: 23 DEC 2009 | DOI: 10.1111/j.1863-2378.2009.01310.x
- R. Vivancos, D. Showell, B. Keeble, S. Goh, M. Kroese, A. Lipp and J. BattersbyArticle first published online: 23 DEC 2009 | DOI: 10.1111/j.1863-2378.2009.01315.x
- M. Scotch, K. Mattocks, P. Rabinowitz and C. BrandtArticle first published online: 16 FEB 2010 | DOI: 10.1111/j.1863-2378.2009.01319.x
- E. K. Leonard, D. L. Pearl, R. L. Finley, N. Janecko, A. S. Peregrine, R. J. Reid-Smith and J. S. WeeseArticle first published online: 16 FEB 2010 | DOI: 10.1111/j.1863-2378.2009.01320.x
- E. Lawaczeck, J. Toporek, J. Cwikla and B. A. MathisonArticle first published online: 12 AUG 2010 | DOI: 10.1111/j.1863-2378.2010.01334.x
Neste Blog realizamos: 1 - Atualização sobre SAÚDE PÚBLICA, PESQUISA BIOMÉDICA E BIOSSEGURANÇA. 2 - Atualização sobre a ocorrência de Doenças de importância em animais de laboratório e outras espécies. 3 - Troca de Informações sobre Doenças Infecciosas, Zoonoses, Licenciamento Ambiental, Defesa Sanitária Animal, Vigilância Sanitária, Boas Práticas de Laboratório e demais assuntos relacionados à sanidade e Saúde Pública.
Pesquisar Neste Blog
quinta-feira, 3 de março de 2011
Estudo genético prevê mutações futuras dos vírus da gripe
 |
| Com o rastreamento das mutações genéticas dos vírus os cientistas poderão, no futuro, se antecipar à própria mutação, fabricando as vacinas antes que as primeiras pessoas comecem a adoecer. |
40 anos de gripe
Cientistas da Universidade da Pensilvânia (EUA) estão decifrando o código genético que mostra qual cepa dos vírus da gripe será predominante em um determinado ano.
Os pesquisadores estão usando supercomputadores e programas especiais de varredura para analisar os genomas ligados às gripes que se espalharam pelo mundo ao longo dos últimos 40 anos.
Os resultados terão implicações para a prontidão da saúde pública global, permitindo que os laboratórios se preparem com as vacinas adequadas a tempo de evitar grandes surtos.
Prevendo mutações futuras
O estudo está permitindo um novo olhar para as mutações dos vírus.
Rastrear mutações isoladas nem sempre é suficiente para entender como o vírus da gripe evolui.
Ao catalogar pares de mudanças genéticas que ocorreram em rápida sucessão, os cientistas descobrem mutações em um dos elementos do par que servem como indicação das mutações que irão ocorrer nos outros.
"Às vezes uma mutação é adaptativa ou funcional somente se ela estiver no contexto de uma certa bagagem genética - isto é, se a proteína já tem alguma outra mutação," explica o Dr. Joshua Plotkin, coautor da pesquisa.
A influência que essas combinações têm sobre a capacidade adaptativa de um organismo é conhecida como epistasia.
"Se você vê uma mutação ocorrer em um Ponto A e, em seguida, vê uma mutação no Ponto B, e esse padrão se repetir, então você tem uma indicação de que A e B se influenciam epistaticamente," disse Plotkin.
"A primeira mutação pode ser inútil isoladamente, mas pode ser uma condição prévia para uma segunda mutação útil [para o organismo]. A primeira mutação é como dar-lhe um prego, e a segunda é como dar-lhe um martelo," explica.
Com isto torna-se possível prever uma mutação antes que ela ocorra de fato, analisando as pré-condições de mutações que ocorreram de fato ao longo das décadas e que beneficiaram os vírus, permitindo que eles atacassem os seres humanos.
Antecipando-se às mutações
Como as mutações geralmente afetam as proteínas superficiais que determinam se o vírus pode entrar e infectar as células humanas, prever quais mutações são mais susceptíveis de ocorrer no futuro próximo tem grandes implicações para a saúde pública.
A produção de vacinas contra a gripe é trabalhosa e demorada, e só pode começar quando há material genético suficiente para isso, ou seja, quando um grande número de pessoas já foram infectadas e não puderam contar com nenhum tratamento ou prevenção.
Com o rastreamento das mutações genéticas dos vírus os cientistas poderão, no futuro, se antecipar à própria mutação, fabricando as vacinas antes que as primeiras pessoas comecem a adoecer.
Postado por
SHDA_CECAL FIOCRUZ
às
07:38
0
comentários

Enviar por e-mailPostar no blog!Compartilhar no XCompartilhar no FacebookCompartilhar com o Pinterest
Satélites artificiais monitoram doenças do espaço
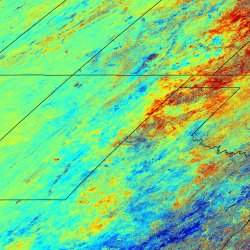 |
A técnica permite a compilação de mapas de risco de doenças, mediante a sobreposição da distribuição dos locais de maior infecção com a distribuição da população humana. |
Hantavirose
O risco de epidemias mortais de hantavirose pode ser avaliado com meses de antecedência usando imagens captadas por satélites artificiais.
A comprovação, que veio por meio do monitoramento em surtos de crescimento na vegetação que induzem um aumento na população de ratos, foi feita por pesquisadores da Universidade de Utah, nos Estados Unidos.
Segundo eles, o método pode ser usado para prever surtos de doenças transmitidas por diversos tipos de roedores e doenças que eles transmitem em todo o mundo.
Doenças transmitidas por ratos
"É uma forma de controlar remotamente uma doença sem ter de sair e capturar animais o tempo todo," disse Denise Dearing, coautora do estudo publicado na revista Global Ecology and Biogeography.
"O satélite mede a intensidade do verde da Terra, e descobrimos que essa intensidade do verde prevê a densidade da população de ratos Peromyscus," explica a bióloga.
Embora o estudo tenha sido focado sobre o hantavírus transmitido pelos ratosPeromyscus, os resultados podem ajudar no combate a outras doenças transmitidas por roedores, como a febre do rato, a doença de Lyme, a peste bubônica, a febre de Lassa, a infecção por salmonela e várias febres hemorrágicas.
O método foi testado em ratos que carregam os hantavírus e se proliferam quando sua fonte de alimento é abundante, "mas pode potencialmente ser aplicado a qualquer animal que responda [ao crescimento da] vegetação," diz Dearing. "Ele teria que ser calibrado para cada uma das espécies específicas de roedores e da doença, mas é realmente poderoso."
Mapas de risco de doenças
O estudo combinou imagens de satélite com dados de milhares de ratos capturados ao longo de três anos na região central de Utah.
O número total de animais capturados e o número de ratos com a doença, uma cepa do hantavírus conhecido como "vírus sem nome" (Sin Nombre virus), ambos subiram após os picos de vegetação detectados pelos satélites.
A técnica permite a compilação de mapas de risco de doenças, mediante a sobreposição da distribuição dos locais de maior população de roedores com a distribuição da população humana.
Postado por
SHDA_CECAL FIOCRUZ
às
07:35
0
comentários

Enviar por e-mailPostar no blog!Compartilhar no XCompartilhar no FacebookCompartilhar com o Pinterest
Mutations Found in Human Induced Pluripotent Stem Cells
ScienceDaily (Mar. 2, 2011) — Ordinary human cells reprogrammed as induced pluripotent stem cells (hiPSCs) may ultimately revolutionize personalized medicine by creating new and diverse therapies unique to individual patients. But important and unanswered questions have persisted about the safety of these cells, in particular whether their genetic material is altered during the reprogramming process.
 |
Image of induced stem cells. |
A new study -- published in the March 3 issue of the journal Natureand led by scientists at the University of California, San Diego in collaboration with other leading stem cell research groups -- finds that the genetic material of reprogrammed cells may in fact be compromised, and suggests that extensive genetic screening of hiPSCs become standard practice before these stem cells are used clinically.
A national team of researchers, co-directed by Kun Zhang, PhD, an assistant professor of bioengineering in the UC San Diego Jacobs School of Engineering, examined 22 different hiPSC lines obtained from seven research groups that employed different methods to reprogram skin cells into pluripotent stem cells. In all of these cell lines, the researchers found protein-coding point mutations, an estimated six mutations per exome. The exome is the part of the genome that contains the genetic instructions for making proteins and other gene products.
"Every single stem cell line we looked at had mutations. Based on our best knowledge, we expected to see 10 times fewer mutations than we actually observed," said Zhang, a faculty member of the Institute for Genomic Medicine and the Institute of Engineering in Medicine, both at UC San Diego.
The findings help answer the question of whether reprogramming adult mammalian cells into hiPSCs affects the overall genome at the fundamental level of single nucleotides. They do. Zhang called the mutations "permanent genome scars."
The scientists said while some of the mutations appeared to be silent, the majority did change specific protein functions, including those in genes associated with causative effects in cancers.
"Reprogrammed stem cells provide an important new tool in the fight against human disease, but to use these cells directly in the clinic, we must ensure that they are safe and that we are able to define their structure and behavior in the most precise terms," said Lawrence S.B. Goldstein, PhD, professor in the Department of Cellular and Molecular Medicine at the UCSD School of Medicine and co-director of the study with Zhang. Goldstein is also director of the UC San Diego Stem Cell Program.
"Our studies open a new window into the genetic behavior of these important types of stem cells and begin to define some new and straightforward safety standards that may help accelerate their use in clinical settings," Goldstein added.
The study examined stem cell lines from many of the leading stem cell research groups in the United States, including lines from the laboratories of James Thomson at the University of Wisconsin-Madison and George Daley at the Children's Hospital Boston, the first U.S.-based labs to reprogram human cells.
"We covered cell lines derived from seven different labs because we wanted to make sure our conclusions are general enough to make realistic extrapolations," said Zhang.
The interdisciplinary team at UC San Diego developed a new, highly sensitive assay to identify mutations that occur at very low frequencies in the starting cells of cell lines. They discovered that roughly half of the mutations found in stem cell lines were present in starting cells at very low levels. That is, they occurred in a few cells sometime during the person's life or during cell culture in the lab, and were somatic or not inherited. The other half of the mutations were too rare to detect in starting cells, meaning they could have occurred during or after reprogramming.
The mutations, which the scientists dubbed "reprogramming-associated mutations," came from three different sources: a first group that included mutations already present in skin cells before reprogramming; a second group of mutations that occurred during reprogramming; and a third group of mutations that occurred after reprogramming, when pluripotent cells began proliferating.
The work is complementary to research published in Cell Stem Cell in January 2011 by another team of scientists at UC San Diego and elsewhere that documented other types of genetic abnormalities in both human embryonic and hiPSC lines after reprogramming and extended culture. That paper reported that human pluripotent and induced pluripotent cells had higher frequencies of genomic aberrations than other cell types. The latest work presents new findings about a different type of important genetic damage: changes occurring during reprogramming in single nucleotides or base pairs that alter the crucial protein building blocks of cells.
"These studies look at two different aspects of stem cell mutations," said Zhang, "but their take-home message is the same -- things can go wrong at the genome level when reprogramming and growing reprogrammed cells. So, to maximize safety, before we put these cells back in the human body for therapeutic purposes, we must be sure that the cells contain the same genome as the recipient, with no cancer-causing or other serious types of mutations."
Additional authors to the paper include Athurva Gore, Zhe Li and Ho-Lim Fung of the Department of Bioengineering, Institute for Genomic Medicine and Institute of Engineering in Medicine, UC San Diego; Jessica E. Young, Isabel Canto, Mason A. Israel and Melissa L. Wilbert, Department of Cellular and Molecular Medicine and Howard Hughes Medical Institute, UC San Diego; Suneet Agarwal, Yuin-Han Loh, Philip D. Manos and George Q. Daley, Division of Pediatric Hematology/Oncology, Children's Hospital Boston and Dana Farber Cancer Institute, Boston; Jessica Antosiewicz-Bourget, Junying Yu and James A. Thomson, Department of Anatomy, University of Wisconsin-Madison; Alessandra Giorgetti, Nuria Montserrat of the Center of Regenerative Medicine, Barcelona, Spain; Juan Carlos Izpisua Belmonte of the Center of Regenerative Medicine, Barcelona, Spain and the Salk Institute for Biological Studies; Evangelos Kiskinis and Kevin Eggan, Howard Hughes Medical Institute, Harvard Stem Cell Institute, Department of Stem Cell and Regenerative Biology, Harvard University; Je-Hyuk Lee, Department of Genetics, Harvard Medical School; Athanasia D. Panopoulos and Sergio Ruiz, Salk Institute for Biological Studies; Ewen F. Kirkness, J. Craig Venter Institute; Derrick J. Rossi, Immune Disease Institute, Children's Hospital Boston.
Postado por
SHDA_CECAL FIOCRUZ
às
07:29
0
comentários

Enviar por e-mailPostar no blog!Compartilhar no XCompartilhar no FacebookCompartilhar com o Pinterest
Key Step in the Development of a Norovirus Treatment
ScienceDaily (Mar. 2, 2011) — With the number of norovirus infection cases rising across the country, scientists from the University of Southampton have successfully crystallised a key norovirus enzyme, which could help in the development of a norovirus treatment.
 |
This is a protein crystal of the Southampton norovirus protease bound to the inhibitor. |
Noroviruses are recognised world-wide as the most important cause of epidemic nonbacterial gastroenteritis (stomach bugs) and pose a significant public health burden, with an estimated one million cases per year in the UK. In the past, noroviruses have also been called 'winter vomiting viruses'.
By crystallising the key protease enzyme, the research team from the University has been able to design an inhibitor that interacts with the enzyme from the 'Southampton' norovirus. The inhibitor works by preventing the enzyme in the norovirus from working, stopping the spread of infection.
The virus is called the Southampton virus because this particular virus was first found in an outbreak that came from a family in Southampton. Traditionally, individual noroviruses are named after the place from which the virus was first found, so for example the very first norovirus is known as Norwalk virus because it discovered in Norwalk in Ohio, America.
University of Southampton virologist Professor Ian Clarke says:
"Noroviruses place a huge burden on the NHS. This is an important step forward in the rational design of new drugs to treat norovirus infections. Now we know the drug works in the test tube, the next step is to see whether we can modify and deliver it to the site where the virus grows."
The research team hopes to translate their laboratory findings into an antiviral treatment for norovirus infection.
The work was performed by research student Rob Hussey in collaboration with University Professor Shoolingin-Jordan, the norovirus research group at Southampton General Hospital and Professor Jon Cooper at University College London. The project was part funded by the University of Southampton, the Hope Charity and the Wellcome Trust.
Postado por
SHDA_CECAL FIOCRUZ
às
07:27
0
comentários

Enviar por e-mailPostar no blog!Compartilhar no XCompartilhar no FacebookCompartilhar com o Pinterest
HIV Vaccine Impacts the Genetic Makeup of the Virus
ScienceDaily (Mar. 2, 2011) — An AIDS vaccine tested in people, but found to be ineffective, influenced the genetic makeup of the virus that slipped past. The findings suggest new ideas for developing HIV vaccines.
The results were published Feb. 27 in Nature Medicine.
This is the first evidence that vaccine-induced cellular immune responses against HIV-1 infection exert selective pressure on the virus. "Selective pressure" refers to environmental demands that favor certain genetic traits over others.
The senior author of the multi-institutional study is Dr. James I. Mullins, University of Washington (UW) professor of microbiology. The research team analyzed the genome sequences in HIV-1 isolated from 68 newly infected volunteers in the STEP HIV-1 vaccine trial. Mullins and the other principal researchers who carried out this study were not involved in the STEP trial.
The STEP trial was a double-blind, Phase 2B test-of-concept of a Merck HIV-1 subtype B vaccine. The vaccine, MRKAd5, was designed to make the body produce infection-fighting white blood cells, commonly called killer T-cells, that could recognize and target specific parts of HIV-1 known as Gag, Pol and Nef.
The STEP trial was conducted at 34 North American, Caribbean, South American and Australian locations where the HIV-1 subtype B was the predominant virus in the local HIV-infected populations. The trial enrolled 3,000 participants.
Preliminary tests indicated the vaccine was encouraging the appearance of the desired virus-attacking cells. More than 75 percent of vaccinated participants produced HIV-1 specific T cells.
Nevertheless, this response to the vaccine did not predict protection. The trial failed. Immunizations were halted, Mullins recalled, after the first interim analysis indicated that the vaccine neither prevented HIV-1 infection nor reduced the load of virus in the body.
"Even though the T-cell responses were not sufficient to prevent infection," Mullins said, "we were interested in whether the vaccine-elicited T-cells had any impact on those strains of HIV-1 that established infections in the study subjects."
The research team tested for a "sieve effect," which, Mullins explained, occurs when a vaccine successfully blocks some strains of virus and not others. The researchers wanted to know, What are the genetic characteristics of those breakthrough viruses that slipped past the immunization barrier erected by the MRKAd5 vaccine?
The research team isolated strains of HIV-1 from both vaccine and placebo recipients in the study, and compared the genetic sequences of the strains. This would help researchers to determine if any changes in the "founder virus" -- the virus first detected in the infection -- might have helped it evade the vaccine-induced immune response and take hold in the vaccinated individuals.
The researchers identified potential T-cell targets in the protein-producing regions of the founder virus genetic sequences and compared these to the virus protein-targets of the vaccine -- Gag, Pol and Nef. The researchers found that the distances for these viral genetic sequences were greater for the viruses taken from the vaccinated individuals, compared to those from the placebo recipients.
The most significant virus genetic site distinguishing vaccine from placebo recipients was in the region known as Gag-84, which was encompassed by several of the viral segments targeted by the vaccine.
Moreover, the researchers said that the extended divergence between the viruses from the vaccinated and the placebo groups was confined only to the sequences for the proteins targeted by the vaccine components (Gag, Pol and Nef) and was not found in other HIV-1 protein sequences. The influence of the vaccine on the virus genotype, Mullins said, was subtle.
Mullins and his team, as well as their collaborators from the STEP trials studies, are doing similar studies of the genetic impact of the Thailand vaccine RV144 on the AIDS virus. The RV144 vaccine was the first to show some probable effectiveness, but its efficacy was not great enough to put it to more general use.
The researchers added that their findings on breakthrough viruses suggest that new vaccines should be designed to put selective pressure on the virus in a controlled manner.
Such a vaccine, Mullins said, should select for genetic mutations in regions of the virus known to be associated with viral control and should avoid selecting for strains that can either escape the immune defense or act as decoys to fool the immune system.
The researchers propose a goal for new designs of vaccines aimed at inducing killer T-cell responses: corner the virus into assuming forms that debilitate it. This would make the infecting virus fitness-impaired -- unable to adapt, reproduce in great numbers and cause disease progression.
"Despite the sad results of the STEP trial," Mullins said, "it has provided clues to ways for science to go forward in the search for an HIV vaccine.
The research was supported by a grant from the U.S. Public Health Service.
In addition to Mullins, others on the research team were: Morgane Rolland, Sodsai Tovanabutra, Allan C. decamp, Nicole Frahm, Peter B. Gilbert, Eric Sanders-Buell, Laura Heath, Craig A. Margaret, Meera Bose, Andrea Bradfield, Annemarie O'Sullivan, Jacqueline Crossler, Teresa Jones, Marty Nau, Kim Wong, Hong Zhoa, Dana N. Raugi, Stephanie Sorensen, Julia Stoddard, Brandon S. Maust, Wenjie Deng, John Hural, Sheri Dubey, Nelson L. Michael, John Shiver, Lawrence Corey, Fusheng Li, Stephen G. Self., Jerome Kim, Susan Buchbinder, Danilo R. Casimiro, Michael N. Robertson, Ann Duerr, M. Juliana McElrath, and Francine E. McCutchan.
Postado por
SHDA_CECAL FIOCRUZ
às
07:25
0
comentários

Enviar por e-mailPostar no blog!Compartilhar no XCompartilhar no FacebookCompartilhar com o Pinterest
Assinar:
Comentários (Atom)

